Q02: Functional microbiota analysis
In project Q02, Thomas Clavel and his team provide expertise in microbiota analysis to the consortium.
Please find detailed information on our protocols and resources on a separate page as follows:
Project Q02 relies on established wet-lab protocols and in-house developed bioinformatic pipelines to offer amplicon and shotgun sequencing services to generate and compare microbiota profiles from all relevant samples collected by the consortium. The project is thereby the basis for harmonized analysis of microbiota profiles for the consortium. This includes the education of young researchers within and outside the consortium on good practices in microbiota analysis by organizing hands-on data analysis workshops.
In addition to sequencing work, to enable translational research by moving from microbiota shifts by sequencing to functional studies with isolates, Q02 relies on our well-established cultivation workflows to isolate specific bacteria. The project also uses a newly established, high-throughput anaerobic platform to create personalized strain collections. Moreover, genetic engineering is implemented to establish causal relationships between target functions from gut bacteria and effects on the host in mouse models.
In summary, Q02 continues to address the needs of CRC projects with respect to microbiota analysis by providing a broad spectrum of standard and advanced cultivation-based, sequencing, and bioinformatic methods.
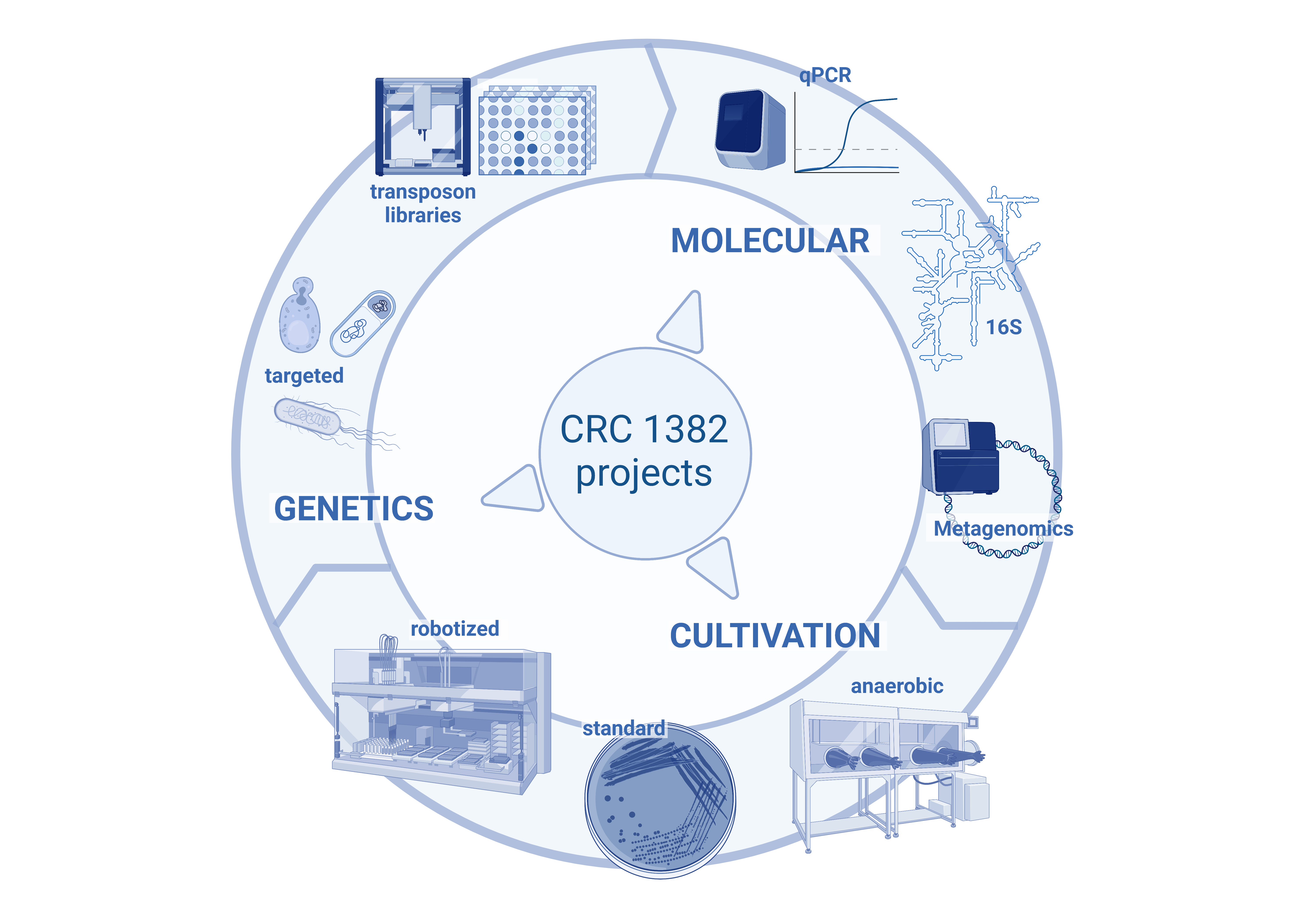
Schematic overview of the work in Q02. The project includes three main arms: (i) Molecular analysis: to provide a flexible portfolio of three main methods to perform diversity, composition, and quantitative analyses adapted to individual CRC project needs; (ii) Cultivation: here we build on our long-lasting experience in this field to enrich the diversity of cultured bacteria for specific CRC projects. This is highly complementary to the work in Aim 1 and enables functional studies in vitro and in vivo. (iii) Microbial genetic engineering: to directly link effects on the host to specific microbial functions based on both, targeted (model organisms) and non-targeted (transposon library) approaches.